MNIST dataset dimensionality reduction example
MNIST dataset dimensionality reduction example
import numpy as np
import matplotlib.pyplot as plt
from sklearn.decomposition import PCA
from sklearn.datasets import fetch_openml
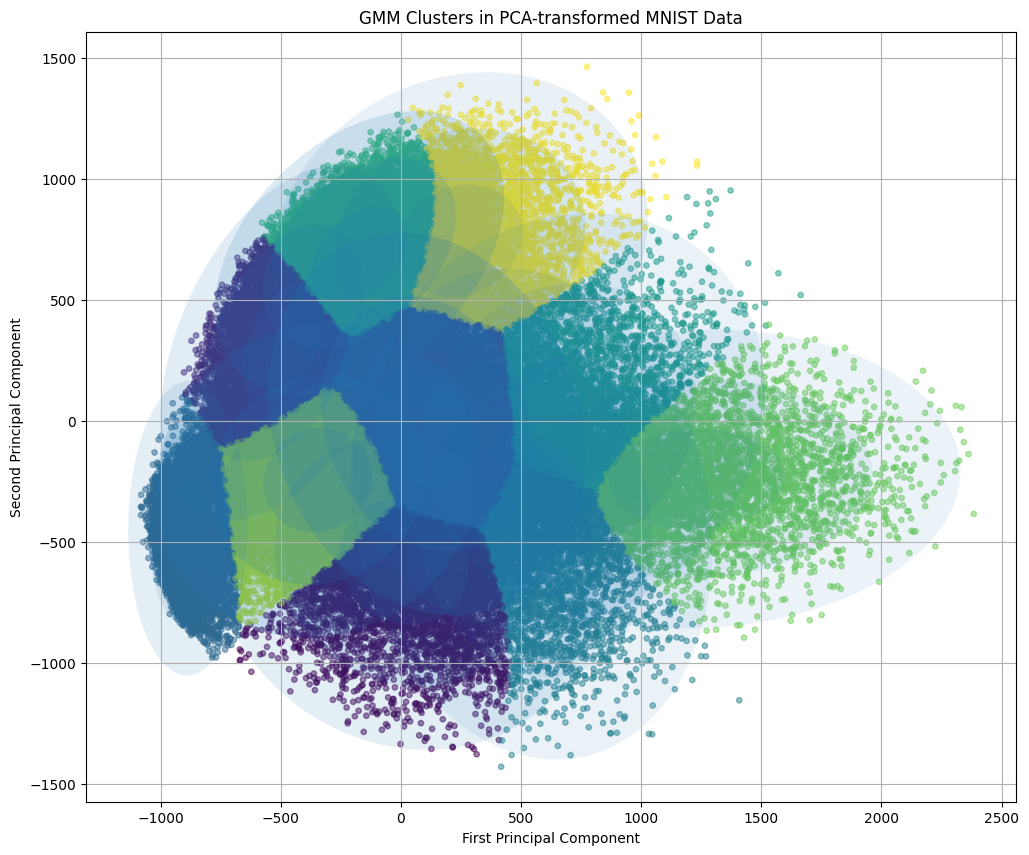
mnist = fetch_openml('mnist_784', version=1, as_frame=False)
X = mnist.data # Feature matrix (70,000 samples of 784 features)
y = mnist.target.astype(int) # Labels (digits 0-9)
# 2. Perform PCA to reduce the dimensionality to 2 components
pca = PCA(n_components=2)
X_pca = pca.fit_transform(X)
# 3. Plot the first two principal components
plt.figure(figsize=(10, 8))
scatter = plt.scatter(X_pca[:, 0], X_pca[:, 1], c=y, cmap='tab10', alpha=0.7, s=15)
plt.xlabel('First Principal Component')
plt.ylabel('Second Principal Component')
plt.title('PCA of MNIST Dataset')
plt.colorbar(scatter, ticks=range(10), label='Digit Label')
plt.grid(True)
plt.show()
from sklearn.mixture import GaussianMixture
# 3. Fit a Gaussian Mixture Model with 10 components
gmm = GaussianMixture(n_components=10, covariance_type='full', random_state=42)
gmm.fit(X_pca)
# 4. Retrieve the means and covariances
means = gmm.means_ # Shape (10, 2)
covariances = gmm.covariances_ # Shape (10, 2, 2)
# 5. Compute standard deviations from covariances
stds = np.sqrt(np.array([np.diag(cov) for cov in covariances])) # Shape (10, 2)
# 6. Print the means and standard deviations
print("Means of the clusters:")
for idx, mean in enumerate(means):
print(f"Cluster {idx}: Mean = {mean}")
print("\nStandard deviations of the clusters:")
for idx, std in enumerate(stds):
print(f"Cluster {idx}: Std Dev = {std}")
# 7. Optional: Plot the GMM clusters with ellipses
def plot_gmm(gmm, X, label=True, ax=None):
import matplotlib as mpl
ax = ax or plt.gca()
labels = gmm.predict(X)
if label:
ax.scatter(X[:, 0], X[:, 1], c=labels, s=15, cmap='viridis', alpha=0.5)
else:
ax.scatter(X[:, 0], X[:, 1], s=15, alpha=0.5)
w_factor = 0.2 / gmm.weights_.max()
for pos, covar, w in zip(gmm.means_, gmm.covariances_, gmm.weights_):
draw_ellipse(pos, covar, alpha=w * w_factor, ax=ax)
def draw_ellipse(position, covariance, ax=None, **kwargs):
from matplotlib.patches import Ellipse
ax = ax or plt.gca()
if covariance.shape == (2, 2):
U, s, Vt = np.linalg.svd(covariance)
angle = np.degrees(np.arctan2(U[1, 0], U[0, 0]))
width, height = 2 * np.sqrt(s)
else:
width, height = 2 * np.sqrt(covariance)
angle = 0
for nsig in range(1, 4): # 1 to 3 standard deviations
ax.add_patch(Ellipse(position, nsig * width, nsig * height,
angle=angle, **kwargs))
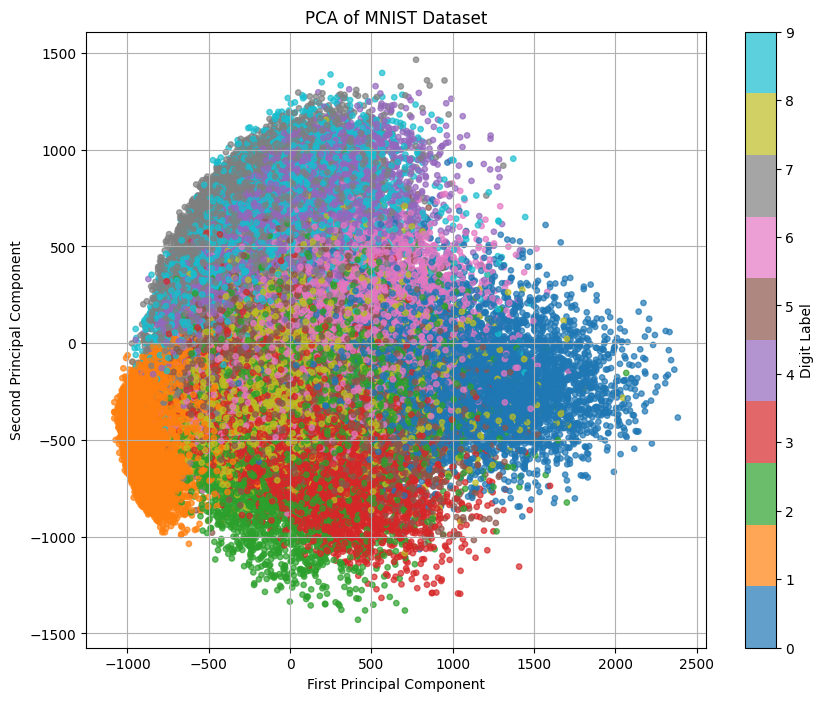
plt.figure(figsize=(12, 10))
plot_gmm(gmm, X_pca)
plt.xlabel('First Principal Component')
plt.ylabel('Second Principal Component')
plt.title('GMM Clusters in PCA-transformed MNIST Data')
plt.grid(True)
plt.show()
Means of the clusters:
Cluster 0: Mean = [ 10.89273388 -582.65412344]
Cluster 1: Mean = [-491.45505023 322.8170719 ]
Cluster 2: Mean = [108.17363694 -11.6977259 ]
Cluster 3: Mean = [-886.40446614 -443.3430705 ]
Cluster 4: Mean = [ 543.55838297 -384.02867352]
Cluster 5: Mean = [657.32338032 -3.80313235]
Cluster 6: Mean = [-172.1257724 692.05765228]
Cluster 7: Mean = [1232.39450602 -239.78532534]
Cluster 8: Mean = [-346.40125825 -244.59018096]
Cluster 9: Mean = [246.66408949 744.93955816]
Standard deviations of the clusters:
Cluster 0: Std Dev = [268.3250472 258.7472777]
Cluster 1: Std Dev = [169.65372386 244.75804164]
Cluster 2: Std Dev = [219.63718487 263.79338739]
Cluster 3: Std Dev = [ 82.56368013 203.04177516]
Cluster 4: Std Dev = [250.9754226 338.35620799]
Cluster 5: Std Dev = [286.11580058 287.72260005]
Cluster 6: Std Dev = [200.71351419 196.54461748]
Cluster 7: Std Dev = [365.46135628 206.15829197]
Cluster 8: Std Dev = [224.42248248 216.4345234 ]
Cluster 9: Std Dev = [260.51560172 232.27217183]